|
Bayesian Analysis of Neutron Star Mass and Radius Observations
|
|
Bayesian Analysis of Neutron Star Mass and Radius Observations
|
Statistical analysis of EOS from M and R constraints. More...
#include <bamr.h>
Public Member Functions | |
| virtual void | run (int argc, char *argv[]) |
| Main wrapper for parsing command-line arguments. | |
Protected Member Functions | |
Main functions called from the command-line interface | |
| virtual int | set_model (std::vector< std::string > &sv, bool itive_com) |
| Set the model for the EOS to use. | |
| virtual int | set_first_point (std::vector< std::string > &sv, bool itive_com) |
| Set the first point in the parameter space. | |
| virtual int | add_data (std::vector< std::string > &sv, bool itive_com) |
| Add a data distribution to the list. | |
| virtual int | mcmc (std::vector< std::string > &sv, bool itive_com) |
| Run a MCMC simulation. More... | |
Protected Attributes | |
| o2scl::err_hnd_cpp | ee |
| Error handler for each thread. | |
| o2scl::table_units | tc |
| Main data table for Markov chain. | |
| size_t | mh_success |
| The number of Metropolis steps which succeeded. | |
| size_t | mh_failure |
| The number of Metropolis steps which failed. | |
| size_t | mcmc_iterations |
| Total number of mcmc iterations. | |
| std::vector< double > | markov_chain |
| Store the full Markov chain. | |
| o2scl::cli | cl |
| Command-line interface. | |
| bool | first_file_update |
| If true, then first_update() has been called. | |
| int | grid_size |
| Number of bins for all histograms (default 100) | |
| size_t | n_chains |
| Number of Markov chain segments. | |
| size_t | chain_size |
| Number of chains. | |
| std::string | model_type |
| A string indicating which model is used, set in set_model(). | |
| bool | has_esym |
| True if the model provides S and L. | |
| bool | has_eos |
| True if the model has an EOS. | |
| size_t | nparams |
| Number of parameters, set in set_model() | |
| double | schwarz_km |
| Schwarzchild radius (set in constructor) | |
| model * | modp |
| The model for the EOS. | |
| model * | modp2 |
| The second copy of the model for the EOS. | |
| o2scl::rng_gsl | gr |
| Random number generator. | |
| std::ofstream | scr_out |
| The screen output file. | |
| std::vector< std::string > | run_args |
| The arguments sent to the command-line. | |
Parameter objects for the 'set' command | |
| o2scl::cli::parameter_double | p_max_time |
| o2scl::cli::parameter_double | p_min_max_mass |
| o2scl::cli::parameter_double | p_step_fac |
| o2scl::cli::parameter_double | p_exit_mass |
| o2scl::cli::parameter_double | p_input_dist_thresh |
| o2scl::cli::parameter_double | p_min_mass |
| o2scl::cli::parameter_int | p_n_warm_up |
| o2scl::cli::parameter_int | p_grid_size |
| o2scl::cli::parameter_int | p_user_seed |
| o2scl::cli::parameter_int | p_max_iters |
| o2scl::cli::parameter_int | p_file_update_iters |
| o2scl::cli::parameter_bool | p_norm_max |
| o2scl::cli::parameter_bool | p_debug_star |
| o2scl::cli::parameter_bool | p_debug_load |
| o2scl::cli::parameter_bool | p_debug_eos |
| o2scl::cli::parameter_bool | p_output_next |
| o2scl::cli::parameter_bool | p_baryon_density |
| o2scl::cli::parameter_bool | p_use_crust |
| o2scl::cli::parameter_bool | p_inc_baryon_mass |
| o2scl::cli::parameter_double | p_nb_low |
| o2scl::cli::parameter_double | p_nb_high |
| o2scl::cli::parameter_double | p_e_low |
| o2scl::cli::parameter_double | p_e_high |
| o2scl::cli::parameter_double | p_m_low |
| o2scl::cli::parameter_double | p_m_high |
Histogram limits | |
| double | nb_low |
| double | nb_high |
| double | e_low |
| double | e_high |
| double | m_low |
| double | m_high |
Grids | |
| o2scl::uniform_grid< double > | nb_grid |
| o2scl::uniform_grid< double > | e_grid |
| o2scl::uniform_grid< double > | m_grid |
Other parameters accessed by 'set' and 'get' | |
| int | file_update_iters |
| The number of MCMC successes between file updates. | |
| bool | debug_load |
| If true, output debug information about the input data files (default false) | |
| bool | norm_max |
| If true, normalize the data distributions so that the max is one, otherwise, normalize so that the integral is one. | |
| int | max_iters |
| Maximum number of iterations (default 0) | |
| bool | use_crust |
| If true, use the default crust (default true) | |
| bool | debug_star |
| If true, output stellar properties for debugging. | |
| bool | debug_eos |
| If true, output equation of state for debugging. | |
| bool | output_next |
| If true, output next point. | |
| bool | baryon_density |
| If true, compute the baryon density. | |
| double | step_fac |
| MCMC stepsize factor (default 15) | |
| double | input_dist_thresh |
| The lower threshold for the input distributions. | |
| double | exit_mass |
| An upper mass threshold. | |
| double | min_mass |
| Minimum mass allowed for any of the individual neutron stars. | |
| double | min_max_mass |
| Minimum allowed maximum mass. | |
| int | n_warm_up |
| Number of warm up steps (successful steps not iterations) More... | |
| double | max_time |
| Time in seconds (3600 seconds is one hour, default is 86400 seconds or 1 day) | |
| int | user_seed |
| If non-zero, use as the seed for the random number generator. | |
| bool | best_detail |
| If true, output more detailed information about the best point. | |
| bool | inc_baryon_mass |
| If true, output information about the baryon mass as well as the gravitational mass. | |
Limits on mass and radius from source data files | |
These are automatically computed in load_mc() as the smallest rectangle in the | |
| double | in_m_min |
| double | in_m_max |
| double | in_r_min |
| double | in_r_max |
MPI properties | |
| int | mpi_rank |
| The MPI processor rank. | |
| int | mpi_nprocs |
| The MPI number of processors. | |
| double | mpi_start_time |
| The MPI starting time. | |
Input neutron star data | |
| std::vector< o2scl::table3d > | source_tables |
| Input probability distributions. | |
| std::vector< std::string > | source_names |
| The names for each source. | |
| std::vector< std::string > | table_names |
| The names of the table in the data file. | |
| std::vector< std::string > | source_fnames |
| File names for each source. | |
| std::vector< std::string > | slice_names |
| Slice names for each source. | |
| size_t | nsources |
| The number of sources. | |
| std::vector< double > | first_mass |
| The initial set of neutron star masses. | |
Parameter limits | |
| entry | low |
| entry | high |
Other variables | |
| ubvector | first_point |
| The first point in the parameter space. | |
| std::string | first_point_file |
| The file containing the initial point. | |
TOV solver objects | |
| o2scl::eos_tov_interp | teos |
| EOS interpolation object for TOV solver. | |
| o2scl::tov_solve * | ts |
| Pointer to a TOV solver. | |
| o2scl::tov_solve * | ts2 |
| Second pointer to a TOV solver. | |
| o2scl::tov_solve | def_ts |
| Default TOV solver. | |
| o2scl::tov_solve | def_ts2 |
| Second default TOV solver. | |
Integer designating how to set the initial point | |
| int | first_point_type |
| static const int | fp_unspecified =-1 |
| static const int | fp_last =-2 |
| static const int | fp_best =-3 |
Internal functions | |
| virtual void | select_mass (entry &e_current, entry &e_next, double mmax) |
| The function which selects the next neutron star masses. | |
| virtual void | table_names_units (std::string &s, std::string &u) |
| Setup column names and units for data table. | |
| virtual int | mcmc_init () |
| Make any necessary preparations for the mcmc() function. More... | |
| virtual bool | make_step (double w_current, double w_next, bool debug, bool warm_up, int iteration) |
| Decide to accept or reject the step. | |
| virtual void | init_grids_table (entry &low, entry &high) |
| Initialize the expectation value objects. More... | |
| virtual void | prepare_eos (entry &e, model &modref, o2scl::tov_solve *tsr, int &success) |
| Further preparations of the EOS before calling the TOV solver. More... | |
| virtual void | add_measurement (entry &e, o2scl::o2_shared_ptr< o2scl::table_units<> >::type tab_eos, o2scl::o2_shared_ptr< o2scl::table_units<> >::type tab_mvsr, double weight, bool new_meas, size_t n_meas, ubvector &weights) |
| Add a measurement. | |
| virtual void | fill_line (entry &e, o2scl::o2_shared_ptr< o2scl::table_units<> >::type tab_eos, o2scl::o2_shared_ptr< o2scl::table_units<> >::type tab_mvsr, double weight, bool new_meas, size_t n_meas, ubvector &weights, std::vector< double > &line) |
Fill vector in line with data from the current Monte Carlo point. | |
| virtual void | first_update (o2scl_hdf::hdf_file &hf, model &modp) |
| Write initial data to HDF file. | |
| virtual void | update_files (std::string fname_prefix, model &modp, entry &e_current) |
Write histogram data to files with prefix fname. | |
| virtual void | setup_cli () |
| Set up the 'cli' object. More... | |
| virtual void | load_mc () |
| Load input probability distributions (called by mcmc()) | |
| virtual double | compute_weight (entry &e, model &modref, o2scl::tov_solve *ts, int &success, ubvector &wgts, bool warm_up) |
Compute 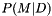 from parameters in entry from parameters in entry e. More... | |
| virtual void | output_best (std::string fname_prefix, entry &e_best, double w_best, o2scl::o2_shared_ptr< o2scl::table_units<> >::type tab_eos, o2scl::o2_shared_ptr< o2scl::table_units<> >::type tab_mvsr, ubvector &wgts) |
| Output the "best" EOS obtained so far (called by mcmc()) | |
| virtual void | compute_star (entry &e, model &modref, o2scl::tov_solve *ts, int &success) |
| Tabulate EOS and then use in cold_nstar. More... | |
|
protectedvirtual |
In order to save the code from copying results over if a step is rejected, the Metropolis steps alternate between two kinds. In the first kind of step, modp is the previous point and modp2 is the new point to be considered, and in the second kind of step, modp2 is the old point and modp is the new point to be considered. This two-step procedure is also why there are two TOV solvers, i.e. ts and ts2.
|
protectedvirtual |
This function is called by mcmc().
|
protectedvirtual |
This function is empty by default.
|
protectedvirtual |
This function just adds the four commands and the 'set' parameters
|
protectedvirtual |
Called by mcmc().
|
virtual |
Called by compute_weight().
|
protected |
Documentation generated with Doxygen. Bamr documentation is under the GNU Free Documentation License.